Counting and mining with the shell
Last updated on 2024-02-23 | Edit this page
Overview
Questions
- How can I count data?
- How can I find data within files?
- How can I combine existing commands to do new things?
Objectives
- Demonstrate counting lines, words, and characters with the shell command wc and appropriate flags
- Use strings to mine files and extract matched lines with the shell
- Create complex single line commands by combining shell commands and regular expressions to mine files
- Redirect a command’s output to a file.
- Process a file instead of keyboard input using redirection.
- Construct command pipelines with two or more stages.
- Explain Unix’s ‘small pieces, loosely joined’ philosophy.
Counting and mining data
Now that you know how to navigate the shell, we will move onto learning how to count and mine data using a few of the standard shell commands. While these commands are unlikely to revolutionise your work by themselves, they’re very versatile and will add to your foundation for working in the shell and for learning to code. The commands also replicate the sorts of uses library users might make of library data.
Counting and sorting
We will begin by counting the contents of files using the Unix shell. We can use the Unix shell to quickly generate counts from across files, something that is tricky to achieve using the graphical user interfaces of standard office suites.
Let’s start by navigating to the directory that contains our data
using the cd command:
Remember, if at any time you are not sure where you are in your
directory structure, use the pwd command to find out:
OUTPUT
/Users/riley/Desktop/shell-lessonAnd let’s just check what files are in the directory and how large
they are with ls -lhS:
OUTPUT
total 139M
-rw-rw-r-- 1 riley staff 126M Jun 10 2015 2014-01_JA.tsv
-rw-r--r-- 1 riley staff 7.4M Jan 31 18:47 2014-01-31_JA-america.tsv
-rw-r--r-- 1 riley staff 3.6M Jan 31 18:47 2014-01-31_JA-africa.tsv
-rw-r--r-- 1 riley staff 1.4M Jan 31 18:47 2014-02-02_JA-britain.tsv
-rw-r--r-- 1 riley staff 598K Jan 31 18:47 gulliver.txt
-rw-r--r-- 1 riley staff 583K Feb 1 22:53 33504-0.txt
drwxr-xr-x 2 riley staff 68 Feb 2 00:58 backupIn this episode we’ll focus on the dataset
2014-01_JA.tsv, that contains journal article metadata, and
the three .tsv files derived from the original dataset.
Each of these three .tsv files includes all data where a
keyword such as africa or america appears in
the ‘Title’ field of 2014-01_JA.tsv.
CSV and TSV Files
CSV (Comma-separated values) is a common plain text format for
storing tabular data, where each record occupies one line and the values
are separated by commas. TSV (Tab-separated values) is just the same
except that values are separated by tabs rather than commas.
Confusingly, CSV is sometimes used to refer to both CSV, TSV and
variations of them. The simplicity of the formats make them great for
exchange and archival. They are not bound to a specific program (unlike
Excel files, say, there is no CSV program, just lots and
lots of programs that support the format, including Excel by the way.),
and you wouldn’t have any problems opening a 40 year old file today if
you came across one.
First, let’s have a look at the largest data file, using the tools we learned in Reading files:
Like 829-0.txt before, the whole dataset cascades by and
can’t really make any sense of that amount of text. To cancel this
on-going concatenation, or indeed any process in the Unix
shell, press Ctrl+C.
In most data files a quick glimpse of the first few lines already tells us a lot about the structure of the dataset, for example the table/column headers:
OUTPUT
File Creator Issue Volume Journal ISSN ID Citation Title Place Labe Language Publisher Date
History_1a-rdf.tsv Doolittle, W. E. 1 59 KIVA -ARIZONA- 0023-1940 (Uk)RN001571862 KIVA -ARIZONA- 59(1), 7-26. (1993) A Method for Distinguishing between Prehistoric and Recent Water and Soil Control Features xxu eng ARIZONA ARCHAEOLOGICAL AND HISTORICAL SOCIETY 1993
History_1a-rdf.tsv Nelson, M. C. 1 59 KIVA -ARIZONA- 0023-1940 (Uk)RN001571874 KIVA -ARIZONA- 59(1), 27-48. (1993) Classic Mimbres Land Use in the Eastern Mimbres Region, Southwestern New Mexico xxu eng ARIZONA ARCHAEOLOGICAL AND HISTORICAL SOCIETY 1993
In the header, we can see the common metadata fields of academic
papers: Creator, Issue, Citation,
etc.
Next, let’s learn about a basic data analysis tool: wc
is the “word count” command: it counts the number of lines, words, and
bytes. Since we love the wildcard operator, let’s run the command
wc *.tsv to get counts for all the .tsv files
in the current directory (it takes a little time to complete):
OUTPUT
13712 511261 3773660 2014-01-31_JA-africa.tsv
27392 1049601 7731914 2014-01-31_JA-america.tsv
507732 17606310 131122144 2014-01_JA.tsv
5375 196999 1453418 2014-02-02_JA-britain.tsv
554211 19364171 144081136 totalThe first three columns contains the number of lines, words and bytes.
If we only have a handful of files to compare, it might be faster or more convenient to just check with Microsoft Excel, OpenRefine or your favourite text editor, but when we have tens, hundreds or thousands of documents, the Unix shell has a clear speed advantage. The real power of the shell comes from being able to combine commands and automate tasks, though. We will touch upon this slightly.
For now, we’ll see how we can build a simple pipeline to find the
shortest file in terms of number of lines. We start by adding the
-l flag to get only the number of lines, not the number of
words and bytes:
OUTPUT
13712 2014-01-31_JA-africa.tsv
27392 2014-01-31_JA-america.tsv
507732 2014-01_JA.tsv
5375 2014-02-02_JA-britain.tsv
554211 totalThe wc command itself doesn’t have a flag to sort the
output, but as we’ll see, we can combine three different shell commands
to get what we want.
First, we have the wc -l *.tsv command. We will save the
output from this command in a new file. To do that, we redirect
the output from the command to a file using the ‘greater than’ sign
(>), like so:
There’s no output now since the output went into the file
lengths.txt, but we can check that the output indeed ended
up in the file using cat or less (or Notepad
or any text editor).
OUTPUT
13712 2014-01-31_JA-africa.tsv
27392 2014-01-31_JA-america.tsv
507732 2014-01_JA.tsv
5375 2014-02-02_JA-britain.tsv
554211 totalNext, there is the sort command. We’ll use the
-n flag to specify that we want numerical sorting, not
lexical sorting, we output the results into yet another file, and we use
cat to check the results:
OUTPUT
5375 2014-02-02_JA-britain.tsv
13712 2014-01-31_JA-africa.tsv
27392 2014-01-31_JA-america.tsv
507732 2014-01_JA.tsv
554211 totalFinally we have our old friend head, that we can use to
get the first line of the sorted-lengths.txt:
OUTPUT
5375 2014-02-02_JA-britain.tsvBut we’re really just interested in the end result, not the
intermediate results now stored in lengths.txt and
sorted-lengths.txt. What if we could send the results from
the first command (wc -l *.tsv) directly to the next
command (sort -n) and then the output from that command to
head -n 1? Luckily we can, using a concept called pipes. On
the command line, you make a pipe with the vertical bar character
|. Let’s try with one pipe first:
OUTPUT
5375 2014-02-02_JA-britain.tsv
13712 2014-01-31_JA-africa.tsv
27392 2014-01-31_JA-america.tsv
507732 2014-01_JA.tsv
554211 totalNotice that this is exactly the same output that ended up in our
sorted-lengths.txt earlier. Let’s add another pipe:
OUTPUT
5375 2014-02-02_JA-britain.tsvIt can take some time to fully grasp pipes and use them efficiently, but it’s a very powerful concept that you will find not only in the shell, but also in most programming languages.
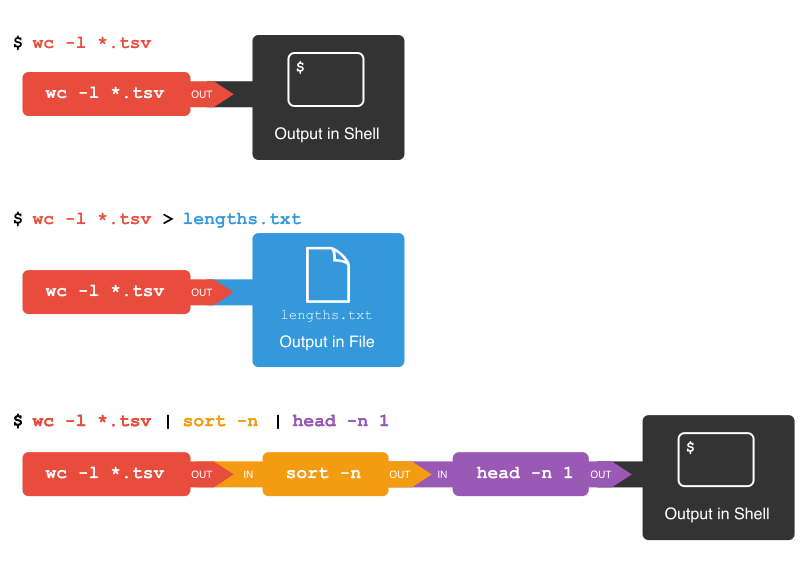
Pipes and Filters
This simple idea is why Unix has been so successful. Instead of
creating enormous programs that try to do many different things, Unix
programmers focus on creating lots of simple tools that each do one job
well, and that work well with each other. This programming model is
called “pipes and filters”. We’ve already seen pipes; a filter is a
program like wc or sort that transforms a
stream of input into a stream of output. Almost all of the standard Unix
tools can work this way: unless told to do otherwise, they read from
standard input, do something with what they’ve read, and write to
standard output.
The key is that any program that reads lines of text from standard input and writes lines of text to standard output can be combined with every other program that behaves this way as well. You can and should write your programs this way so that you and other people can put those programs into pipes to multiply their power.
Count the number of words, sort and print (faded example)
To count the total lines in every tsv file, sort the
results and then print the first line of the file we use the
following:
Now let’s change the scenario. We want to know the 10 files that
contain the most words. Check the manual for the
wc command (either using man wc or
wc --help) to see if you can find out what flag to use to
print out the number of words (but not the number of lines and bytes).
Fill in the blanks below to count the words for each file, put them into
order, and then make an output of the 10 files with the most words
(Hint: The sort command sorts in ascending order by default).
From man wc, you will see that there is a
-w flag to print the number of words:
OUTPUT
-w The number of words in each input file is written to the standard
output.So to print the word counts of the .tsv files:
OUTPUT
511261 2014-01-31_JA-africa.tsv
1049601 2014-01-31_JA-america.tsv
17606310 2014-01_JA.tsv
196999 2014-02-02_JA-britain.tsv
19364171 totalAnd to sort the lines numerically:
OUTPUT
196999 2014-02-02_JA-britain.tsv
511261 2014-01-31_JA-africa.tsv
1049601 2014-01-31_JA-america.tsv
17606310 2014-01_JA.tsv
19364171 totalMining or searching
Searching for something in one or more files is something we’ll often
need to do, so let’s introduce a command for doing that:
grep (short for global regular expression
print). As the name suggests, it supports regular expressions
and is therefore only limited by your imagination, the shape of your
data, and - when working with thousands or millions of files - the
processing power at your disposal.
To begin using grep, first navigate to the
shell-lesson directory if not already there. Then create a
new directory “results”:
Now let’s try our first search:
Remember that the shell will expand *.tsv to a list of
all the .tsv files in the directory. grep will
then search these for instances of the string “1999” and print the
matching lines.
Press the up arrow once in order to cycle back to your most recent
action. Amend grep 1999 *.tsv to
grep -c 1999 *.tsv and press enter.
OUTPUT
2014-01-31_JA-africa.tsv:804
2014-01-31_JA-america.tsv:1478
2014-01_JA.tsv:28767
2014-02-02_JA-britain.tsv:284The shell now prints the number of times the string 1999 appeared in each file. If you look at the output from the previous command, this tends to refer to the date field for each journal article.
We will try another search:
OUTPUT
2014-01-31_JA-africa.tsv:20
2014-01-31_JA-america.tsv:34
2014-01_JA.tsv:867
2014-02-02_JA-britain.tsv:9We got back the counts of the instances of the string
revolution within the files. Now, amend the above command
to the below and observe how the output of each is different:
OUTPUT
2014-01-31_JA-africa.tsv:118
2014-01-31_JA-america.tsv:1018
2014-01_JA.tsv:9327
2014-02-02_JA-britain.tsv:122This repeats the query, but prints a case insensitive count
(including instances of both revolution and
Revolution and other variants). Note how the count has
increased nearly 30 fold for those journal article titles that contain
the keyword ‘america’. As before, cycling back and adding
> results/, followed by a filename (ideally in .txt
format), will save the results to a data file.
So far we have counted strings in files and printed to the shell or
to file those counts. But the real power of grep comes in
that you can also use it to create subsets of tabulated data (or indeed
any data) from one or multiple files.
This script looks in the defined files and prints any lines
containing revolution (without regard to case) to the
shell. We let the shell add today’s date to the filename:
This saves the subsetted data to a new file.
However, if we look at this file, it contains every instance of the
string ‘revolution’ including as a single word and as part of other
words such as ‘revolutionary’. This perhaps isn’t as useful as we
thought… Thankfully, the -w flag instructs
grep to look for whole words only, giving us greater
precision in our search.
This script looks in both of the defined files and exports any lines
containing the whole word revolution (without regard to
case) to the specified .tsv file.
We can show the difference between the files we created.
OUTPUT
10585 2016-07-19_JAi-revolution.tsv
7779 2016-07-19_JAiw-revolution.tsv
18364 totalUsing date --help (on Git Bash for Windows or Linux) or
man date (on macOS or Linux) will show you that the
+ option introduces a date format, where %Y,
%m and %d are replaced by the year, month, and
day respectively. There are many other percent-codes you could use.
You might also see that -I is short for --iso-8601, which
essentially avoids the confusion between the European and American date
formats DD.MM.YYYY and MM/DD/YYYY.
Finally, we’ll use the regular expression syntax covered earlier to search for similar words.
Basic, extended, and PERL-compatible regular expressions
There are, unfortunately, different
ways of writing regular expressions. Across its various versions,
grep supports “basic”, at least two types of “extended”,
and “PERL-compatible” regular expressions. This is a common cause of
confusion, since most tutorials, including ours, teach regular
expressions compatible with the PERL programming language, but
grep uses basic by default. Unless you want to remember the
details, make your life easy by always using the most advanced regular
expressions your version of grep supports (-E
flag on macOS X, -P on most other platforms) or when doing
something more complex than searching for a plain string.
The regular expression ‘fr[ae]nc[eh]’ will match “france”, “french”, but also “frence” and “franch”. It’s generally a good idea to enclose the expression in single quotation marks, since that ensures the shell sends it directly to grep without any processing (such as trying to expand the wildcard operator *).
The shell will print out each matching line.
We include the -o flag to print only the matching part
of the lines e.g. (handy for isolating/checking results):
Pair up with your neighbor and work on these exercises:
Searching with regular expressions
Use regular expressions to find all ISSN numbers (four digits
followed by hyphen followed by four digits) in
2014-01_JA.tsv and print the results to a file
results/issns.tsv. Note that you might have to use the
-E flag (or -P with some versions of
grep, e.g. with Git Bash on Windows).
or
It is worth checking the file to make sure grep has
interpreted the pattern correctly. You could use the less
command for this.
The -o flag means that only the ISSN itself is printed
out, instead of the whole line.
If you came up with something more advanced, perhaps including word boundaries, please share your result in the collaborative document and give yourself a pat on the shoulder.
Finding unique values
If you pipe something to the uniq command, it will
filter out adjacent duplicate lines. In order for the ‘uniq’ command to
only return unique values though, it needs to be used with the ‘sort’
command. Try piping the output from the command in the last exercise to
sort and then piping these results to ‘uniq’ and then
wc -l to count the number of unique ISSN values.
Using a Loop to Count Words
We will now use a loop to automate the counting of certain words
within a document. For this, we will be using the Little
Women e-book from Project
Gutenberg. The file is inside the shell-lesson folder
and named pg514.txt. Let’s rename the file to
littlewomen.txt.
$ mv pg514.txt littlewomen.txtThis renames the file to something easier to remember.
Now let’s create our loop. In the loop, we will ask the computer to go through the text, looking for each girl’s name, and count the number of times it appears. The results will print to the screen.
BASH
$ for name in "Jo" "Meg" "Beth" "Amy"
> do
> echo "$name"
> grep -wo "$name" littlewomen.txt | wc -l
> doneOUTPUT
Jo
1355
Meg
683
Beth
459
Amy
645What is happening in the loop?
-
echo "$name"is printing the current value of$name -
grep "$name" littlewomen.txtfinds each line that contains the value stored in$name. The-wflag finds only the whole word that is the value stored in$nameand the-oflag pulls this value out from the line it is in to give you the actual words to count as lines in themselves. - The output from the
grepcommand is redirected with the pipe,|(without the pipe and the rest of the line, the output fromgrepwould print directly to the screen) -
wc -lcounts the number of lines (because we used the-lflag) sent fromgrep. Becausegreponly returned lines that contained the value stored in$name,wc -lcorresponds to the number of occurrences of each girl’s name.
Why are the variables double-quoted here?
In episode 4 we learned to use
"$..."as a safeguard against white-space being misinterpreted. Why could we omit the"-quotes in the above example?What happens if you add
"Louisa May Alcott"to the first line of the loop and remove the"from$namein the loop’s code?
Because we are explicitly listing the names after
in, and those contain no white-space. However, for consistency it’s better to use rather once too often than once too rarely.Without
"-quoting$name, the last loop will try to executegrep Louisa May Alcott littlewomen.txt.grepinterprets only the first word as the search pattern, butMayandAlcottas filenames. This produces two errors and a possibly untrustworthy count:
Selecting columns from our article dataset
When you receive data it will often contain more columns or variables
than you need for your work. If you want to select only the columns you
need for your analysis, you can use the cut command to do
so. cut is a tool for extracting sections from a file. For
instance, say we want to retain only the Creator,
Volume, Journal, and Citation
columns from our article data. With cut we’d:
OUTPUT
Creator Volume Journal Citation
Doolittle, W. E. 59 KIVA -ARIZONA- KIVA -ARIZONA- 59(1), 7-26. (1993)
Nelson, M. C. 59 KIVA -ARIZONA- KIVA -ARIZONA- 59(1), 27-48. (1993)
Deegan, A. C. 59 KIVA -ARIZONA- KIVA -ARIZONA- 59(1), 49-64. (1993)
Stone, T. 59 KIVA -ARIZONA- KIVA -ARIZONA- 59(1), 65-82. (1993)
Adams, W. Y. 1 NORTHEAST AFRICAN STUDIES NORTHEAST AFRICAN STUDIES 1(2/3), 7-18. (1994)
Beswick, S. F. 1 NORTHEAST AFRICAN STUDIES NORTHEAST AFRICAN STUDIES 1(2/3), 19-48. (1994)
Cheeseboro, A. Q. 1 NORTHEAST AFRICAN STUDIES NORTHEAST AFRICAN STUDIES 1(2/3), 49-74. (1994)
Duany, W. 1 NORTHEAST AFRICAN STUDIES NORTHEAST AFRICAN STUDIES 1(2/3), 75-102. (1994)
Mohamed Ibrahim Khalil 1 NORTHEAST AFRICAN STUDIES NORTHEAST AFRICAN STUDIES 1(2/3), 103-118. (1994)Above we used cut and the -f flag to
indicate which columns we want to retain. cut works on tab
delimited files by default. We can use the flag -d to
change this to a comma, or semicolon or another delimiter. If you are
unsure of your column position and the file has headers on the first
line, we can use head -n 1 <filename> to print those
out.
First, let’s see where our desired columns are:
OUTPUT
File Creator Issue Volume Journal ISSN ID Citation Title Place Labe Language Publisher DateOk, now we know Issue is column 3, Volume
4, Language 11, and Publisher 12. We use these
positional column numbers to construct our cut command:
cut -f 3,4,11,12 2014-01_JA.tsv > 2014-01_JA_ivlp.tsvWe can confirm this worked by running head on the file:
head 2014-01_JA_ivlp.tsvOUTPUT
Issue Volume Language Publisher
1 59 eng ARIZONA ARCHAEOLOGICAL AND HISTORICAL SOCIETY
1 59 eng ARIZONA ARCHAEOLOGICAL AND HISTORICAL SOCIETY
1 59 eng ARIZONA ARCHAEOLOGICAL AND HISTORICAL SOCIETY
1 59 eng ARIZONA ARCHAEOLOGICAL AND HISTORICAL SOCIETY